So far we have explained what we mean by reproducible research and explained some of the additional benefits.
In this section we cover some of the (real and perceived) barriers you may face in making your work reproducible.

Figure 1:A slide outlining some of the barriers to reproducible research from Kirstie Whitaker’s talk about The Turing Way at csv,conf,v4 in May 2019. Used under a CC-BY 4.0 license. DOI: Whitaker (2019).
This chapter outlines some of those barriers, and a few suggestions to get around them. The barriers to reproducible research can be described in three main groups. The first, and hardest to overcome are those relating to the current incentive structure in academic research: Limited incentives to give evidence against yourself (or “Plead the fifth”), the known publication bias towards novel findings, the fact that reproducible or open research may be held to higher standards than others, and that all this effort is not considered for promotion. Then there are the technical and theoretical challenges of working with big data and complex computational infrastructure and remembering that being reproducible does not mean the answer is right. We finish with three barriers considering the pressures on individual data scientists: that this work can be perceived to take extra time, that you may be required to support additional users (spoiler: you aren’t!), and that you and members of your team might require additional skills. The good news is that helping you learn those skills is exactly what The Turing Way is here for!
Limited incentives to give evidence against yourself¶
The Fifth Amendment to the United States Constitution includes a clause that no one “shall be compelled in any criminal case to be a witness against themselves”. (Edited to gender-neutral language.) To “plead the fifth” means that someone chooses not to give evidence that there might have been something wrong in their past behaviour. They have the right to remain silent.
We know that no one wants to incriminate themselves, and also that no one is infallible. Putting your code and data online can be very revealing and intimidating, and it is part of the human condition to be nervous of being judged by others. Although there is no law governing the communication of reproducible research - unless you commit explicit fraud in your work - sharing errors that you find in your work is heavily disincentivised.

Figure 2:An illustration of the “plead the fifth” barrier where our current culture disincentivises acknowledging and correcting mistakes. Illustration by The Ludic Group LLP from Kirstie Whitaker’s keynote presentation at Scientific Data in 2017. Used under a CC-BY 4.0 license. DOI: Scientific Data et al. (2017).
Giving evidence against yourself, particularly if you find mistakes in published material, is difficult and stressful. But we need to balance that individual cost against the fact that releasing code can help other researchers provide feedback, learn and may help them in their research. In fact, you will almost certainly find that publishing your code and data documentation motivates you to conduct your analyses to a higher standard. Being careful about what you write down, and documenting your decisions, can also help generate new ideas for yourself and for others.
Most importantly, we need to move away from a culture where publishing nothing is safer than publishing something. The Turing Way is here to help you take little steps towards being more reproducible as your career progresses. We don’t want anyone to feel alone, or “not good enough” as they start and continue their open research journey.
Publication bias towards novel findings¶
Novel results are not necessarily accurate or interesting but they are rewarded in the academic world! Papers that do not find statistically significant relationships are hard to publish, particularly if the results do not reproduce previously published findings. (That includes statistically significant findings that go in the opposite direction to already published work.) Similarly, an article might be less likely to be accepted to a journal or a conference if it successfully reproduces already-published results instead of producing a new set. There’s a good chance that reviewers will say “we already know this” and reject the submission.
The bias towards novelty in data science means many researchers are disincentivised from doing the work to document, test and share their code and data. John Ioannidis published an influential paper in 2005 titled “Why Most Published Research Findings Are False” Ioannidis, 2005 which discusses the many factors that contribute to publication bias. Given these biases, it is very likely that there is a lot of duplicated work in data science. Too many different researchers are asking the same question, not getting the answer they expect or want, and then not telling anyone what they have found.
This barrier is not specific to computational reproducibility as we define it in The Turing Way. However, it is a major cultural barrier to transparent communication, and affects project design. The Turing Way community are advocating in all the places we are able, for the systemic culture change that is required to dismantle the current publication and academic credit biases towards novelty over rigour.
Held to higher standards than others¶
A researcher who makes their work reproducible by sharing their code and data may be held to a higher standard than other researchers. If authors share nothing at all, then all readers of a manuscript or conference paper can do is trust (or not trust) the results.
If code and data are available, peer reviewers may go looking for differences in the implementation. They may come back with new ideas on ways to analyse the data because they have been able to experiment with the work. There is a risk that they then require additional changes from the authors of the submitted manuscript before it is accepted for peer review.
As we described in the "Plead the Fifth" section above, the solution to this challenge is to align career incentives so that doing what is best for science also benefits the individuals involved.
Not considered for promotion¶
In the current academic system, a primary consideration for promotion is the proven ability to be awarded grants and recruit students. Both funding bodies and prospective students value novelty and this behaviour is reflected in preferentially rewarding papers with a high journal impact factor. It is likely part of the human condition to be motivated by things that are new or surprising, but as discussed above, this bias towards novelty causes a systematic publication bias.
More broadly, the promotion system in academia tends to reward individuals who have shown themselves to be different from others in their field. That means sharing code and data to make it easy for “competitors” to do the same work ends up being discouraged by promotion and funding selection panels. A good example of this bias is the Nobel Prize award which only goes to a small number of researchers each year, and as such “overlooks many of its important contributors” (Ed Yong, The Atlantic, 2017). One of the goals of The Turing Way is to draw attention to the misalignment of the tenure and promotion process with collaborative and reproducible data science.
Big data and complex computational infrastructure¶
Big data is conceptualised in different ways by different researchers. “Big” data may be complex, come from a variety of data sources, is large in storage volume and/or be streamed at very high temporal resolution. Although there are ways to set random seeds and take snapshots of a dataset at a particular moment in time, it can be difficult to have identical data across different runs of an analysis pipeline. This is particularly relevant in the context of tools for parallel computing. For example, some data such as flight tracking or internet traffic is so big that it can not be stored and must be processed as it is streamed in real time.
A more common challenge for “big data” researchers is the variability of software performance across operating systems and how quickly the tools change over time. An almost constantly changing ecosystem of data science technologies is available, which means reproducing results in the future is highly variable and dependent on using perfectly backwards compatible tools as they develop. Very often the results of statistical tests will vary depending on the configuration of the infrastructure that was used in each of the experiments, making it very hard to independently reproduce a result. Experiments are often dependent on random initialisation for iterative algorithms and not all software includes the ability to fix a pseudorandom number without limiting parallelisation capabilities (for example in Tensorflow). These tools can require in depth technical skills which are not widely available to data scientists. The Apache Hadoop framework, for instance, is extremely complex to deploy data science experiments without strong software and hardware engineering knowledge.
Even “standard” high performance computing, can be difficult to set up to be perfectly reproducible, particularly across different cloud computing providers or institutional configurations. The Turing Way contains chapters to help data scientists learn skills in reproducible computational environments including containers such as docker and ways to version control your software libraries. We are always open to more contributions as the technology to support reproducible research in very large datasets or for complex modelling evolves.
Being reproducible does not mean the answer is right¶
By making the code and data used to produce a result openly available to others, our results may be reproduced but mistakes made by the initial author can be carried through. Getting the same wrong answer each time is a step in the right direction, but still very much a wrong answer!
This barrier isn’t really a barrier to reproducible research as much as a caveat that investing time in reproducibility doesn’t necessarily mean that you’re doing better science. You can consider computational reproducibility as being necessary but not sufficient for high quality research. A critical approach is needed, rather than naively using existing software or implementing statistical methods without understanding what they do. See, for example, a discussion in August 2019 about whether the default settings for Scikit-learn’s implementation of logistic regression are misleading to new users. Interpretability and interoperability are required to properly evaluate the original research and to strengthen findings.
Takes time¶
Making an analysis reproducible takes time and effort, particularly at the start of the project. This may include agreeing upon a testing framework, setting up version control such as a Github repository and continuous integration, and managing data. Throughout the project, time may be required to maintain the reproducible pipeline.
Time may also be spent communicating with collaborators to agree on which parts of the project may be open source and when and how these outputs are shared. Researchers may find that they need to “upskill” their colleagues to allow the team to benefit from reproducibility tools such as git and GitHub, containers, Jupyter notebooks, or databases.
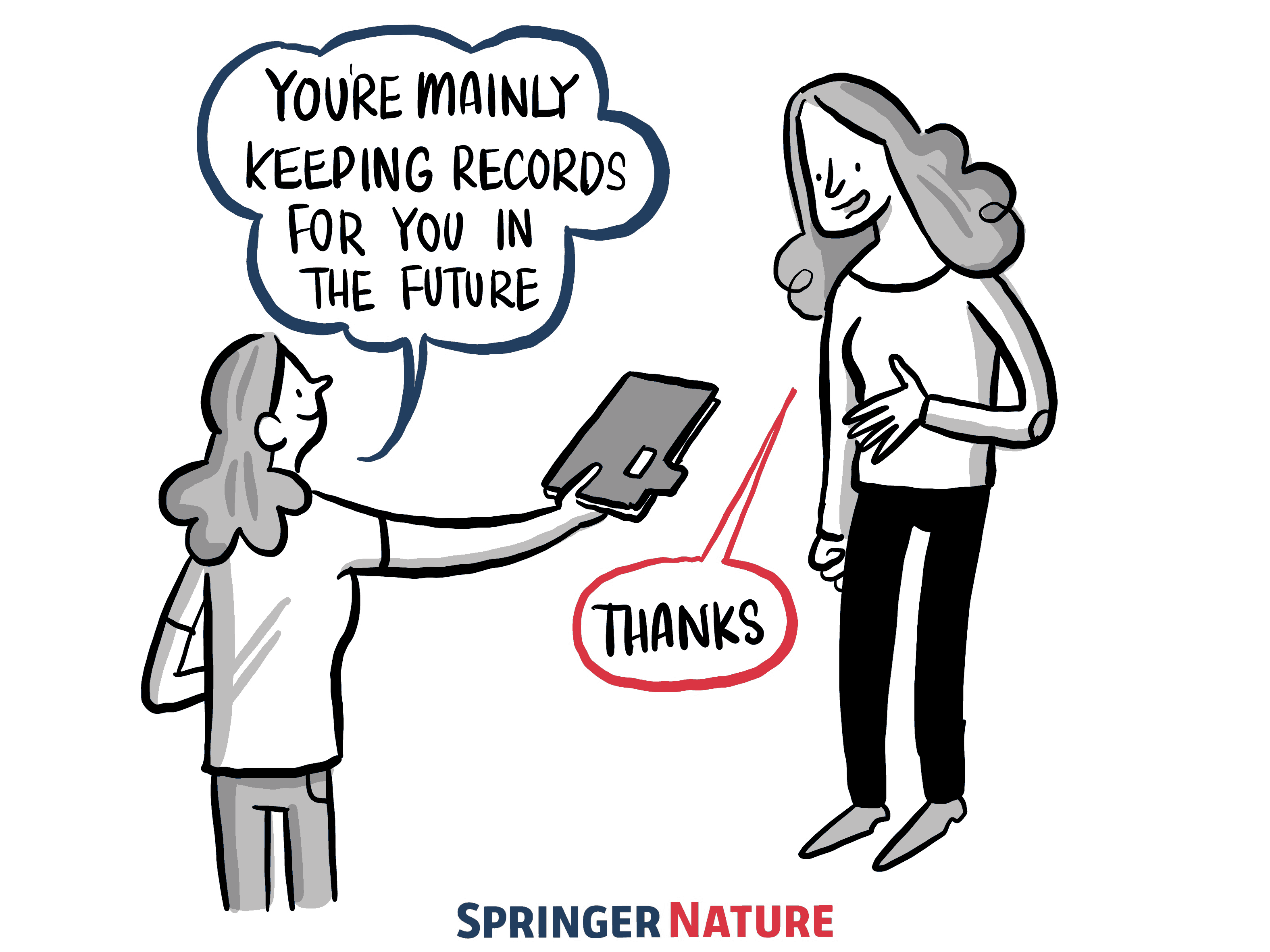
Figure 3:Although making clear documentation may feel like it is taking a lot of time at the moment, you are helping yourself and your collaborators remember what you have done so it is easy to reuse the work or make changes in the future. Illustration by The Ludic Group LLP from Kirstie Whitaker’s keynote presentation at Scientific Data in 2017. Used under a CC-BY 4.0 license. DOI: Scientific Data et al. (2017).
However, The Turing Way community advocates that this time is more than made up for by the end of the project. Take as a thought experiment a reviewer asking for “just one more analysis” when the publication has been submitted to a journal. In many cases, this request will come 6 to 12 months after the research team have worked with the raw data. It can be very hard to go back in time to find the one part of the pipeline that the reviewer has asked you to change. If the work is fully reproducible, including version-controlled data and figure generating code, this analysis will be very fast to run and incorporate into the final research output. The analysis pipeline can be easily adapted as needed in response to co-author and reviewer requests. It can also be easily reused for future research projects.
Support additional users¶
Many people worry that by making their analysis reproducible they will be required to answer lots of questions from future users of their code. These questions may cover software incompatibility across operating systems and the dependencies changing over time (see the Big data and complex computational infrastructure barrier above). They may also include questions about how to adjust the code for a different purpose.
This barrier is based in part on conflating “reproducible” with “open” research. The Turing Way definition of "reproducible" doesn’t require authors to support the expansion and re-use of the data and code beyond running the exact analyses that generate the published results in the accompanying manuscript.
In almost all cases, making code and data open source requires better documentation than a researcher would write for themselves. This can feel like an additional barrier, although - as discussed in the previous section on reproducible research taking extra time it is likely that the primary beneficiaries of well commented and tested code with detailed documentation are the research team - particularly the principal investigator of the project - themselves.
Requires additional skills¶
As you can tell from the ever-growing number of chapters in The Turing Way, working reproducibly requires skills that aren’t always taught in training programmes. You - or someone in your team - might need to develop expertise in data engineering, research software engineering, technical writing for documentation or project management on GitHub. That is a major barrier when the current incentive structures are not aligned with learning these skills (see the barriers on plead the fifth, publication bias towards novel findings, held to higher standards than others, and not considered for a promotion!) However, this is the primary barrier that we at The Turing Way are working to dismantle with you. We hope you enjoy learning these skills with us and that you’ll help us to improve the book as you do.
“A journey of a thousand miles begins with a single step” (Chinese philosopher Lao Tzu).
We hope that by working towards helping you learn some of these valuable skills we also dismantle some of the more structural barriers to reproducible research.
Further reading and additional resources¶
You can watch Kirstie Whitaker describe some of these barriers in her talk about The Turing Way at csv,conf,v4 in May 2019. You can use and re-use her slides under a CC-BY licence via Zenodo (doi: Whitaker (2019)). The section describing the slide below starts around 5 minutes into the video.
- Whitaker, K. (2019). The Turing Way: Sharing the responsibility of reproducibility. 10.5281/ZENODO.2669547
- Ioannidis, J. P. A. (2005). Why Most Published Research Findings Are False. PLOS Medicine, 2(8), null. 10.1371/journal.pmed.0020124